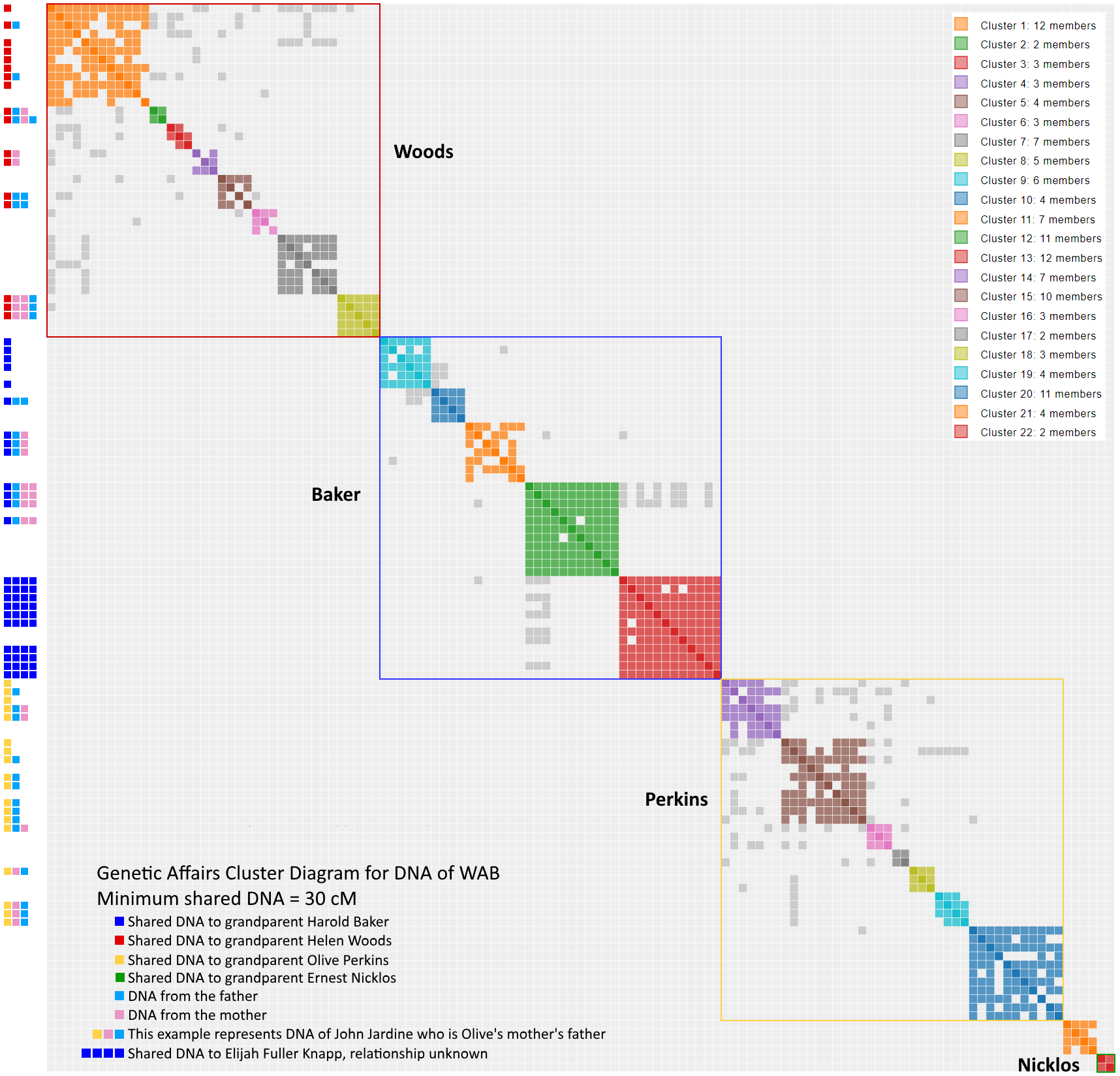
Part 1 introduced the Genetic Affairs program which organizes the various ancestors by creating clusters of DNA shared with WAB. As mentioned, any shared match above 30 cM suggests a likely genealogical match. By the end of the article, clusters 20, 21 and 22 remained unidentified, and there were a series of nine results with > 30 cM that have only one match (making them a lonely cluster of one). Now comes the slow process of examining each of these twelve clusters one-by-one. The hope is to understand each one, and identify its place in the tree, with emphasis on uncovering the Nicklos subgroup, consisting of Ernest Nicholas, Charlotte Zieryacks, James McDowell of Scotland (not to be confused with James McDowell of Pennsylvania in the Woods line) and Isabella McLeod.
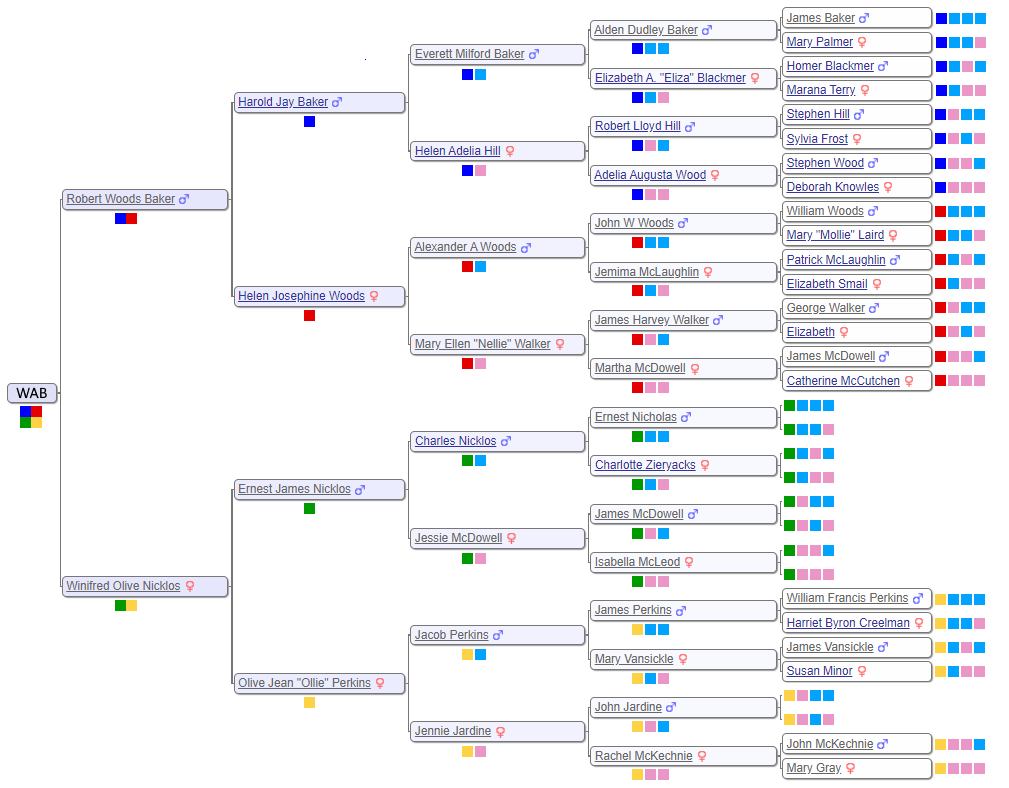
There are a lot of different ways to investigate each cluster. Some people have elaborate trees that we can examine for possible matches. Others have snippets of a tree which we can expand by building “quick and dirty” trees. We can look for additional shared matches of a 30 cM individual down to 20 cM. It’s important to include the geographic clues since we know where WAB’s ancestors lived and when they lived there. We can write messages to the individuals to see if they want to collaborate, although the response rate is frustratingly low. Ultimately, we need a matching family tree that intersects WAB’s.
Extend-a-group
Here’s a nice trick to allow analysis of a subgroup. AncestryDNA allows us to assign shared matches to one or more custom groups. So I converted the Woods, Baker and Perkins subgroups into ancestryDNA custom groups. Genetic Affairs can be configured to run these custom groups. Furthermore, we can set it to extend the group to look for all shared match to a lower threshold (hence, my pet name of “extend-a-group”). When I use this technique for the Baker subgroup of 41 matches, I get a list of around 200 shared Baker related matches down to 20 cM. Then I go back to Ancestry, and assign these 200 to a new group called “Baker to 20 cM”. It takes a little bit of art to massage this list. Matches that are too closely related might be removed. Matches that Genetic Affairs missed might get added. Then I run Genetic affairs again (with extend feature unchecked). Viola! We have a cluster diagram for our deep dive in the Baker, Perkins or Woods lines.
The Cryptocluster and the Pseudocluster
Despite all these efforts some clusters remain unidentified. These clusters contain a lot of shared DNA and robust trees, but with no connection found to WAB. I am going to coin the term “cryptocluster” because of the way these clusters hold their secrets. Cryptoclusters might be legitimate or they might be fake earning them my derisive term “pseudocluster”. I don’t like pseudoclusters because they do not lead to a common ancestor, yet they suck up a lot of research time. Currently, I can think of five reasons why a cluster remains a mystery:
- Potential Causes of Legit Cryptoclusters
- I am wrong – The result points to an erroneous or unknown branch with my tree, possibly due to a NPE (see below)
- They are wrong – Other members of the cluster may be presenting inaccurate information
- Everyone is wrong – The underlying genealogical record is faulty.
- Endogamy – Marriage of somewhat related individuals (see below)
- Potential Causes of Pseudoclusters
- A false positive – The result in an individual match is wrong due to random variation
- Pile up – The result of a group of matches is wrong due to a systemic issue in the DNA (see below)
- A faulty computer algorithm – Genetic Affairs or Ancestry has created a cluster in error
The Nonpaternity Event (NPE)
Sometimes a cryptocluster points to a completely unexpected direction: the surprise DNA result. Genealogists have dubbed these discoveries as nonpaternity events (NPE’s). It’s such a nondescript term. I have heard many NPEs better described as “shenanigans”. Because they occur for a variety of reasons (adoption, out-of-wedlock births, etc), the surprise unmasks a messy side to DNA testing. Without getting into an ethical argument of uncovering these family secrets, it should be noted that it can be very difficult to prove the results with certainty because the supporting genealogy evidence has been swept under the family rug. WAB’s DNA has uncovered a few NPE’s, all over 100 years old. Fortunately, the major branches of the tree has survived intact (so far). If the amount of shared DNA is high enough, I can usually figure what likely happened. However, if the event happened too long ago, then the amount of shared DNA will get too low to unravel the story. The surprise result will remain a cryptocluster.
I have identified a special subset of cryptocluster. The sequence NNNN in cluster 13 of the Genetic Affairs chart points a guy named Elijah Fuller Knapp. Who is that?, or as a Louisiana Cajun might ask, whodat? While he could be a NPE, other possibilities exist. For the time being, cluster 13 is a whodat cryptocluster.
Endogamy
Think of endogamy as “cousin love”. Not the first cousin stereotype. Instead, consider second cousins, third cousins, second half cousins, etc. Let us say you live in a small, isolated community and you want to marry someone who shares your interests, values and culture. That special someone could also easily share a recent common ancestor. The effect on DNA results can be profound. You can see it in the cluster results in two significant ways. First, you will see high levels of shared matches in endogamous results, since shared DNA comes from multiples sources. A 30 cM match from a endogamous branch appears to be more closely related than it is. Stated another way, endogamy acts as a shared match magnifier. Second, you will see a whole bunch of the gray dots in Genetic Affairs since there are multiple common ancestors in a given cluster. Look no further than my father’s side if you want an extreme example of endogamy. Clusters 1 to 21 map the DNA of my paternal grandfather, born of Ashkenasi Jewish parents, a classic endogamous population. You have to look hard to find cluster 22 representing the DNA of my paternal grandmother. “Cluster Bomb” would seem the appropriate term.
Since WAB’s grandparents arrived from such different geographic locales, much of WAB’s tree would seem devoid of endogamy. Or is it? Take a look at all the gray inside the boxes of the Woods, Baker and Perkins subgroups.
Pile Up
Sometimes sharing is detected among an impossibly high number of relations. If you look at the genomes on the chromosomes, particular regions appear to “pile up”. From my limited scope of understanding, everybody has unique areas of pile up on their DNA genome. You really need a chromosome map to identify them. Unfortunately, we are limited to ancestryDNA’s view of the world. AncestryDNA knows about pile up, and its algorithm tries to exclude known problematic regions when it determines shared matches. To me, the odds are low that this algorithm is 100% effective in weeding out pile up.
| Confidence Score | Approximate amount of shared cM’s | Likelihood of a single recent common ancestor |
|---|---|---|
| Extremely high | > 60 cMs | Virtually 100% |
| Very High | 45 – 60 cMs | About 99% |
| High | 30 – 45 cMs | About 95% |
| Good | 16 – 30 cMs | About 50% |
| Moderate | 6 – 16 cMs | 15 – 50% |
Now, let us revisit the ancestryDNA odds table where it states “the likelihood of a single recent common ancestor” is 50% for a 16-30 cM match. Normally, we think that these odds apply to a shared match of one person to another. Pile up suggests that these odds are not necessarily random. Instead, shared match errors may be correlated (the same error happens repeatedly in a given group). As a result, the confidence score could apply to a whole cluster, or part of a cluster. I have tried to use 20 cM as a cutoff to avoid pile up or any other cause of a pseudocluster, but it may not be good enough. If the error is so correlated, then it might be that 50% of entire clusters are pseudoclusters when Genetic Affairs is run at the lower 20 cM threshold.
What does a pile up look like? Rick Marshall, writing in the Genetic Affairs User Group, used chromosome maps to identify a pile up region in his DNA. He then showed the results on Genetic Affairs. I have seen similar results in my cluster diagrams as illustrated below. Whereas we can think of endogamy as a shared match magnifier, pile up acts as a shared match parasite. Here are the notable characteristics:
- The cluster is really big compared to the clusters around it. DNA is piling up.
- The cluster may contain a few legitimate matches in the upper left corner representing the legit host cluster. Those matches have the highest amount of shared DNA. The rest of the cluster is the parasitic pile up DNA latching onto the host.
- You wouldn’t find any common ancestors, because none exist.
- Because it has no matches, the pile up region has few grays dots.
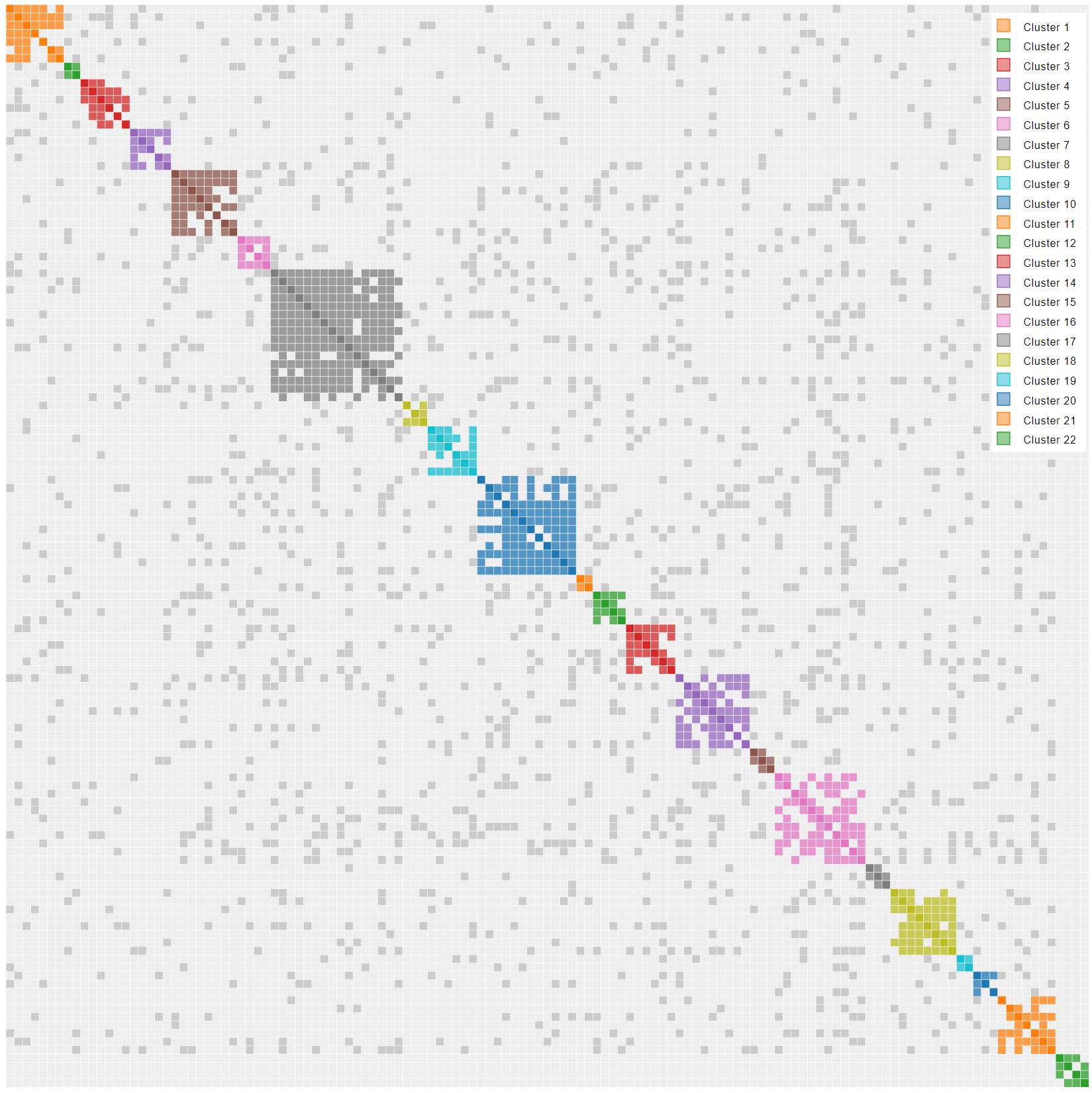
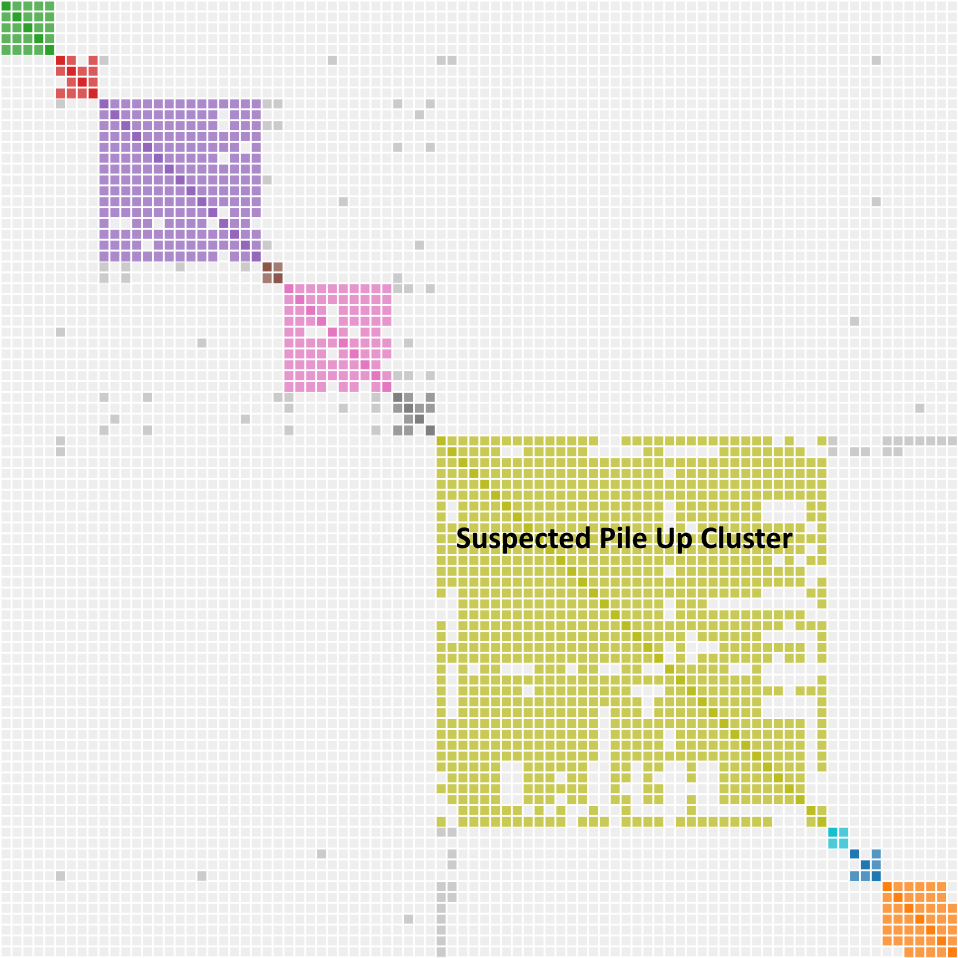
Many of the comments above are observational with a dash of pure science to give me cover. More observations and testing will be necessary to understand the cluster charts at lower thresholds. You have to be flexible in this analysis since new tools, techniques and people get added all the time. Recently, a new single match has surfaced. As you will see, single match 10 has proven to be extremely important.
Cluster 20: This cluster was omitted from the Perkins subgroup despite a single gray dot that intersects. So the extend-a-cluster technique was run without this cluster. Now, when I compare my results to the Perkins at the 20 cM threshold, I see 4-5 shared matches – all to the Perkins subgroup. Cluster 20 has become a cryptocluster within the Perkins subgroup.
Click here to see cluster 20 results
| Cluster 20 Match Name at 30 cM | Shared Matches at 20 cM | Match | Found on a Extend-a-cluster list? |
|---|---|---|---|
| Erica Plyler | 51.1 | Perkins | |
| William Manuel | 30.3 | Perkins | |
| Esther Cameron | 24.1 | Perkins | |
| Michael Johnson | 24.3 | Perkins | |
| vickimagee_1 | 20.2 | Perkins | |
| H.C. by aiuzzolino1 | 40.1 | ||
| Esther Cameron | 24.1 | Perkins | |
| Michael Johnson | 24.3 | Perkins | |
| vickimagee_1 | 24.3 | Perkins | |
| Todd Rank | 40.1 | ||
| Ester Cameron | 24.1 | Perkins | |
| Jane Westergaard-Nimocks | 24.3 | Perkins | |
| Nicky_C | 37.1 | ||
| Esther Cameron | 24.1 | Perkins | |
| Michael Johnson | 24.3 | Perkins | |
| vickimagee_1 | 20.2 | Perkins | |
| T.U. by Mary Lou Upton | 33.1 | Not Found | |
| Lacy Ayers | 33.3 | Not Found | |
| Peyton Gifford | 32.1 | Not Found | |
| jdliz82 | 32.1 | Not Found | |
| James Blake | 31.1 | Not Found | |
| Jane Westergaard-Nimocks | Perkins | ||
| Nancy French | 31.1 | ||
| Esther Cameron | Perkins | ||
| Muriel Bissell | 30.2 | ||
| Esther Cameron | Perkins |
Cluster 21: Four people in the cluster, two with trees and no obvious overlap with WAB tree. Many people in the trees have upstate New York connections, which might suggest a Baker connection, but nothing yet. No connection to DNA of Baker, Perkins or Woods subgroups down to 20 cM.
Click here to see cluster 21 results
| Match Name | Shared Match to 20 cM | Length of Match | Comments |
|---|---|---|---|
| Barbara Mellor | 43.2 | No | |
| C.S by Alisa Mayer | 38.2 | ||
| ginapayne761 | 38.2 | ||
| nancyandnelly | 35.1 | ||
| Gayle Lauriano | 27.2 | ||
| C.S. by Alisa Mayer | 38.2 | Click here for tree. | |
| Barbara Mellor | 43.2 | ||
| ginapayne761 | 38.2 | ||
| JohnPeper | 25.1 | ||
| Amanda Fletcher | 23.1 | ||
| Gordon Ellis | 23.1 | ||
| Deborah Ellis | 22.1 | ||
| William Fletcher | 21.1 | ||
| ginapayne761 | 38 cM / 2 segs | Click here for tree. | |
| Barbara Mellor | 43.2 | ||
| C.S by Alisa Mayer | 38.2 | ||
| nancyandnelly | 35.1 | ||
| JohnPeper | 25.1 | ||
| Amanda Fletcher | 23.1 | ||
| RSmaldon | 23.1 | ||
| William Fletcher | 21.1 | ||
| Timothy Hampton | 21.2 | ||
| nancyandnelly | 35 cm / 1 seg | No | |
| Barbara Mellor | 43.2 | ||
| ginapayne761 | 38.2 |
Cluster 22: Very interesting cluster. There should be an obvious relation because the top match, LC, shares an impressive 77 cM of DNA. He also has posted an well-researched tree to cross-reference. Unfortunately, no common ancestor here. The second member of the cluster, EW, shares 58 cM, but has no tree available. I was, however, able to create a tree for another shared match of 29 cM, JJ. Again, no common ancestor. However, JJ has one ancestor named Elsie Slotz who was born Dec 1873 in Saxony Germany. So I wandering if she might be related to our Ernest Nicholas line who also came from Saxony. Much more work to do on this one.
Click here to see cluster 22 results
| 30 cM Match Name | Shared Name to 20 cM | Length of Match | Comments |
|---|---|---|---|
| LC | 77.6 | Click for tree | |
| EW | 58.4 | ||
| JJ | 29.3 | Click for tree | |
| EW | 58.4 | No | |
| LC | 77.6 |
Single Match 1: G.G. managed by mgilbreth62 (53 cM / 2 segs)
G.G. shares 53 cM of DNA with WAB. Based on mgilbreth62’s snippet of the family tree, GG is son of Beatrice I (Goodridge) Gilbreth born near Rochester, NY. Breatrice was also part of my Casa de Schwartz tree, making our common ancestors, Stephen and Sylvia (Frost) Hill. WAB and GG are likely 4th cousins.
Click here to see single match 1 results
| Single Match Name | Shared Match to 20 cM | Length of Match | Comments |
|---|---|---|---|
| G.G. by mgilbreth62 | 53 cM / 2 segs | Yes. Click here. | |
| Leslie Notham | Common Ancestor: Stephen & Sylvia (Frost) Hill. | ||
| Makenna Adams | |||
| L.F. by 1_rafstamps |
Single Match 2: Loretta Penning (49 cM / 4 segs)
Loretta does not have a usable tree, but she does have two shared matches, both of whom are clearly associated with the Woods family line. So Loretta’s DNA match will be thrown into the “Woods down to 30 cM” pot for further analysis.
Click here to see single match 2 results
| 30 cM Match Name Name | Shared Match to 20 cM | Length of Match | Comments |
|---|---|---|---|
| Loretta Penning | 49.4 | Tree unusable | |
| C.V by PeggyADK | 218.12 | Common ancestor is William Woods & Mary Laird | |
| D.F. by jkfulk1619 | 23.1 | ||
| Mike Fulkerson | 22.2 | ||
| dtarango1 | 20.2 | Common ancestor is Patrick McLaughlin & Elizabeth Smail |
Single Match 3: Craig Terkelsen (40 cM / 4 segs)
Some hope for this match. This group may provide a link to the McDowell family who married in the the Nicklos line. See “Elizabeth McDowell” born 1835 in Ayr Scotland in the tree of DarthTorment
Click here to see single match 3 results
| Shared Match Name | Length of Match | Comments |
|---|---|---|
| DarthTorment | 22.2 | Click to see tree |
| G.N. by deborah mccray | 21.1 | No Tree |
| Craig Campbell | 21.1 | No Tree |
Single Match 4: Connie Peabody (38 cM / 2 segs)
Connie’s has a tree and it does not intersect with WAB’s. However, one of the shared matches has a common relation of Homer and Marana (Terry) Blackmer, making it likely to be part of the the Baker subgroup
Click here to see single match 4 results
| Match Name | Shared Matches to 20 cM | Match | Comments |
|---|---|---|---|
| Connie Peabody | 38.2 | Click here to see tree. | |
| krichmond1963 | 37.3 | Common ancestor, Homer & Marana (Terry) Blackmer. Click here see. | |
| Russell Finch | 29.1 | ||
| Elizabeth Reedy | 28.2 | Tree unusable. |
Single Match 5: W.W. managed by Patty Bueker (37 cM / 3 segs)
No tree for WW. No connection yet. Seven shared matches at 20 cM / 1 segment level, suggesting that these members may be part of a pile up region.
Click here to see single match 5 results
| Match Name | Shared Match to 20 cM | Match | Comments |
|---|---|---|---|
| W.W. managed by Patty Bueker | 37.3 | No | |
| F.C. by Beverly Linquist | 24.1 | Click here to see tree. | |
| M.M. by mmandbs | 20.1 | ||
| G.D. by Kathryn Frasier | 20.1 | ||
| Richard Deese by sideese9047 | 20.1 | ||
| Eric Oden | 20.1 | ||
| Amanda Roth | 20.1 | ||
| C.H. by Genealellie | 20.1 | Private Tree | |
| S.W. by Genealellie | 20.1 | Private Tree |
Single Match 6: P.H. managed by Aggie Henry (35 cM / 2 segs)
Shared matches down to 20 cM share DNA with the Perkins subgroup. The trees of several of the individual intersect the Jacob and Katherine Minor.
Click here to see single match 6 results
| Match Name | Shared Matches down to 20 cM | Length of Match | Comments |
|---|---|---|---|
| P.H. managed by Aggie Henry | 35.2 | No Tree | |
| Stacey Brown | 31.1 | ||
| barryandrobin_1 | 29.3 | shares DNA with Perkins subgroup | |
| sandylefebvre by Julie Lefebvre | 27.2 | shares DNA with Perkins subgroup | |
| jlefebvre163 by Julie Lefebvre | 27.2 | shares DNA with Perkins subgroup | |
| bonjohnpc | 23.2 | common ancestor Jacob & Katherine Minor | |
| M.G. by jmhird1 | 22.1 | common ancestor Jacob & Katherine Minor | |
| Bill Daniels | 21.1 | shares DNA with Perkins subgroup | |
| jennifer kilty | 21.1 | shares DNA with Perkins subgroup | |
| lindaberry441 | 20.1 | common ancestor Jacob & Katherine Minor |
Single Match 7: Miriam Mathews (34 cM / 2 segs)
All matches shared common ancestor of John & Agnes (Lossee) McLaughlin, making it a connection to the Woods subgroup.
Click here to see single match 7 results
| Match Name | Length of Match | Tree? | |
|---|---|---|---|
| Miriam Mathews | 34.2 | Common ancestor: John & Agnes McLaughlin. Click here to see. | |
| C.V. by PeggyADK | 218.12 | Common ancestor: William Woods & Mary Laird | |
| C.D. by PeggyADK | 49.7 | Common ancestor is William Woods & Mary Laird | |
| cydemmons | 25.7 | Common ancestor is John & Agnes McLaughlin |
Single Match 8: ScotterMac (32 cM / 2 segs)
This cluster needs more work. No connection to DNA of Baker, Perkins or Woods subgroups down to 20 cM. Only a single tree with no overlap to WAB’s tree.
Click here to see single match 8 results
| Match Name | Shared Matches to 20 cM | Length of Match | Comments |
|---|---|---|---|
| ScotterMac | 34 cM / 2 segs | Tree Unusable | |
| Susan E Acevedo | 29.2 | Click to see tree | |
| km39497 | 29.3 | Private Tree | |
| D.C. by alefteye | 24.2 | No Tree |
Single Match 9: Lisa Rusyn (31 cM / 2 segs)
Lisa shares 31 cM and has a tree also. Various ancestors connected to Pennsylvania, Ontario and upstate New York. Unfortunately, the connection remains elusive
Click here to see single match 9 results
| Match Name | Shared Matches to 20 cM | Match | Tree? |
|---|---|---|---|
| Lisa Rusyn | 31.2 | Yes. Click here. | |
| Danielle Greene | 29.2 | ||
| Olivia Austin | 28.2 | ||
| Bob Rudesill | 28.1 | ||
| T.S. by Katherine Manolakas | 26.2 |
Single Match 10: Joan Lankton (97 cM / 4 segs)
Better to be lucky than good. Joan has a shared match to DarthTorment of Single Match 3. She also has a shared match with JL who can be found on WAB’s family tree. Therefore single matches 3 and 10 connect to the McDowell line in the Nicklos subgroup.
Click here to see single match 10 results
| Match Name | Length of Match (cM) | Useful Tree? | ||
|---|---|---|---|---|
| WAB | JD | JN | ||
| JL | 23 | 117 | 72 | Yes |
| JLankton | 97 | 21 | 107 | Yes |
| SLankton | None | 12 | 23 | Yes |
| libertyandersonsweeny | 25 | 19 | 26 | No |
| DarthTorment | 22 | None | 27 | Yes |
| bsinc204 | 15 | 20 | None | Yes |
Conclusion
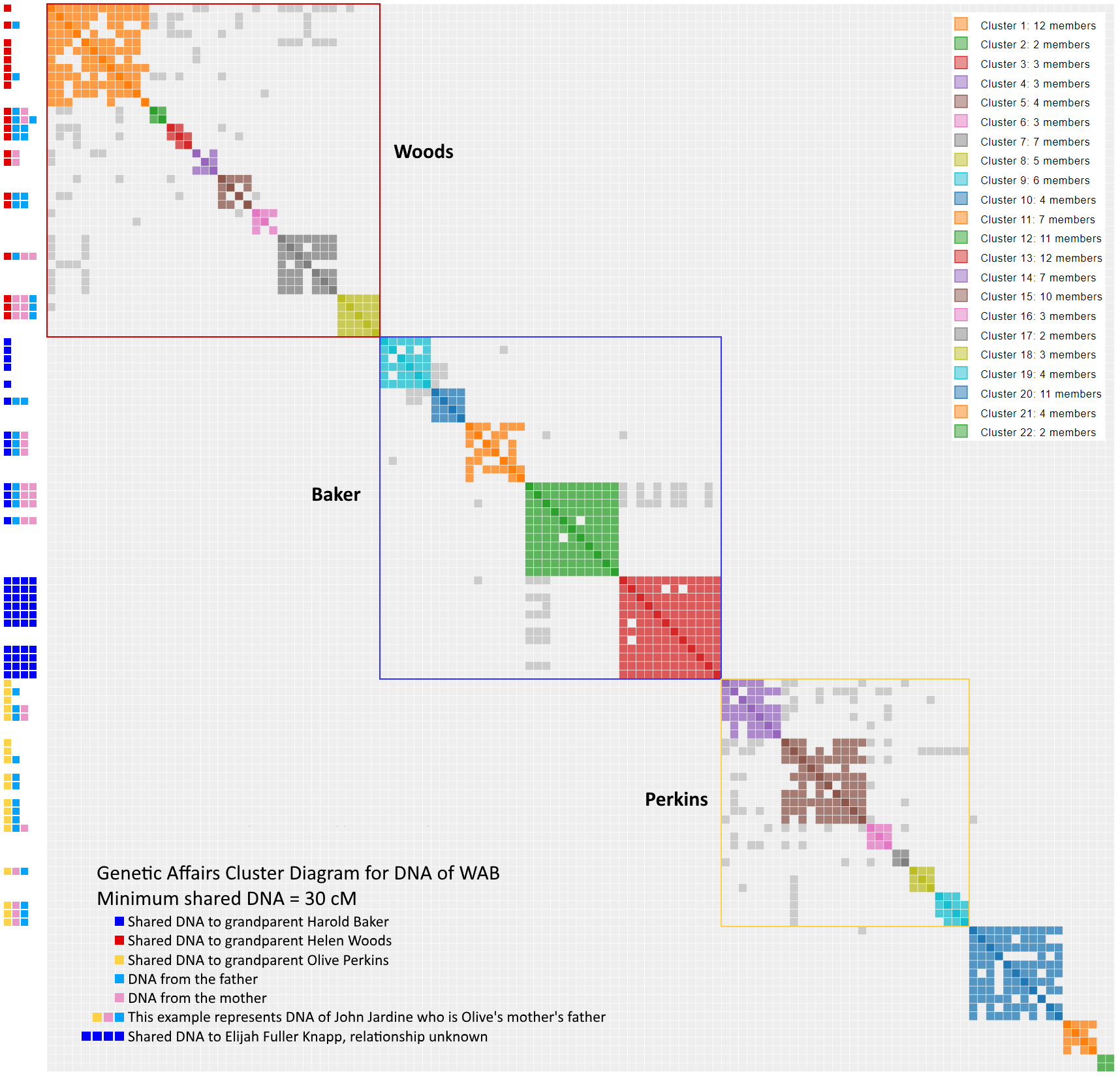
It looks like we succeeded in our goal to identify possible individuals associated with WAB’s Nicklos branch with cluster 22 and single matches 3 & 10. The cluster chart below was updated to reflect our new understanding of WAB’s DNA. There remains some unanswered matches that may yield still yield some surprises (Cluster 21 and four single matches). Despite all the talk of pseudoclusters, these results at 30 cM should have a legit cryptocluster answers. This page will be updated as progress gets made.